Segmentation tutorials¶
GUI¶
Watch this video tutorial:
Or follow the steps below:
After opening the VentMapp3r GUI, click “Ventricle” under the “Segmentation” tab. Wait for a new pop-up window to appear. The window should look like the image below.
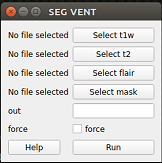 ventricle pop up window
ventricle pop up window
Click “Select t1w” and chose your T1 image. Click “Run”. Click either or both “Select t2w” and “Select flair” (with respect to the sequences you have) and select the correspending images. Type your desired output name in the “out” box. Check the box next to “force” if you want to overwrite a previous output. Click “Run”. Your output file will automatically appear in your t1w folder.
Command Line¶
ventmapper seg_vent
Optional arguments:
-h, --help show this help message and exit
-s , --subj input subject
-fl , --flair input Flair
-t1 , --t1w input T1-weighted
-t2 , --t2w input T2-weighted
-m , --mask brain mask
-o , --out output prediction
-f, --force overwrite existing segmentation
-ss , --session input session for longitudinal studies
Examples:
ventmapper seg_vent -s subjectname -b
ventmapper seg_vent -t1 subject_T1_nu.nii.gz -o subject_vent.nii.gz
The output should look like this.:
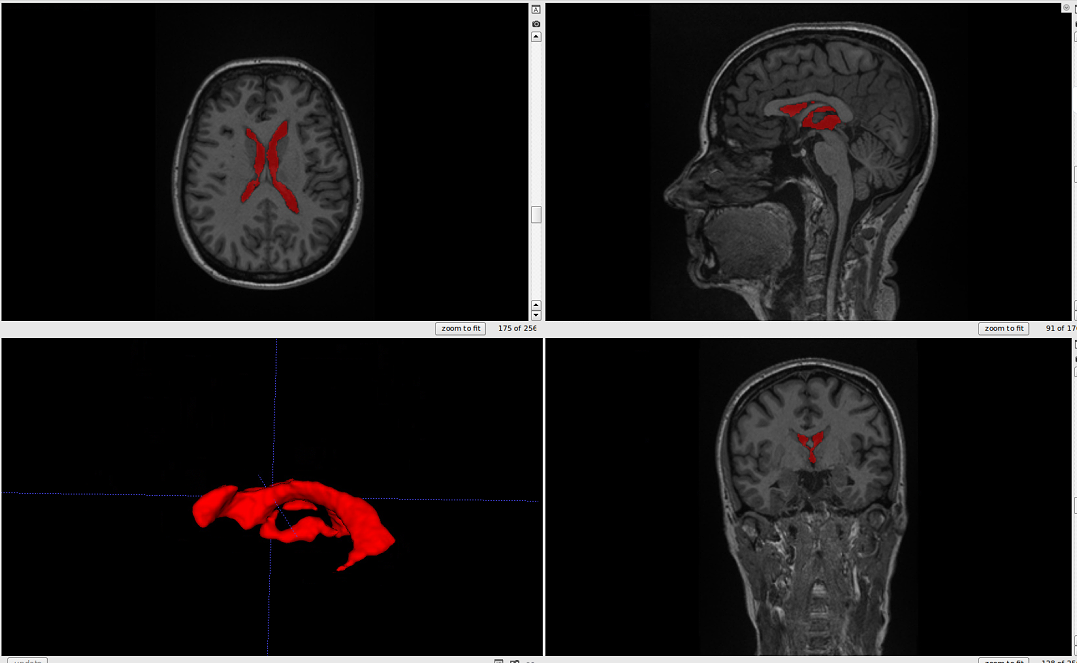 ventricle segmentation
ventricle segmentation
 IMAGE ALT TEXT
IMAGE ALT TEXT